Feature selection using random forest
Examples
library(dplyr)
meta_dat1 <- t(meta_dat) %>%
as.data.frame() %>%
dplyr::mutate(group=group)
result_ML_RF <- ML_RF(meta_dat1)
result_ML_RF$p
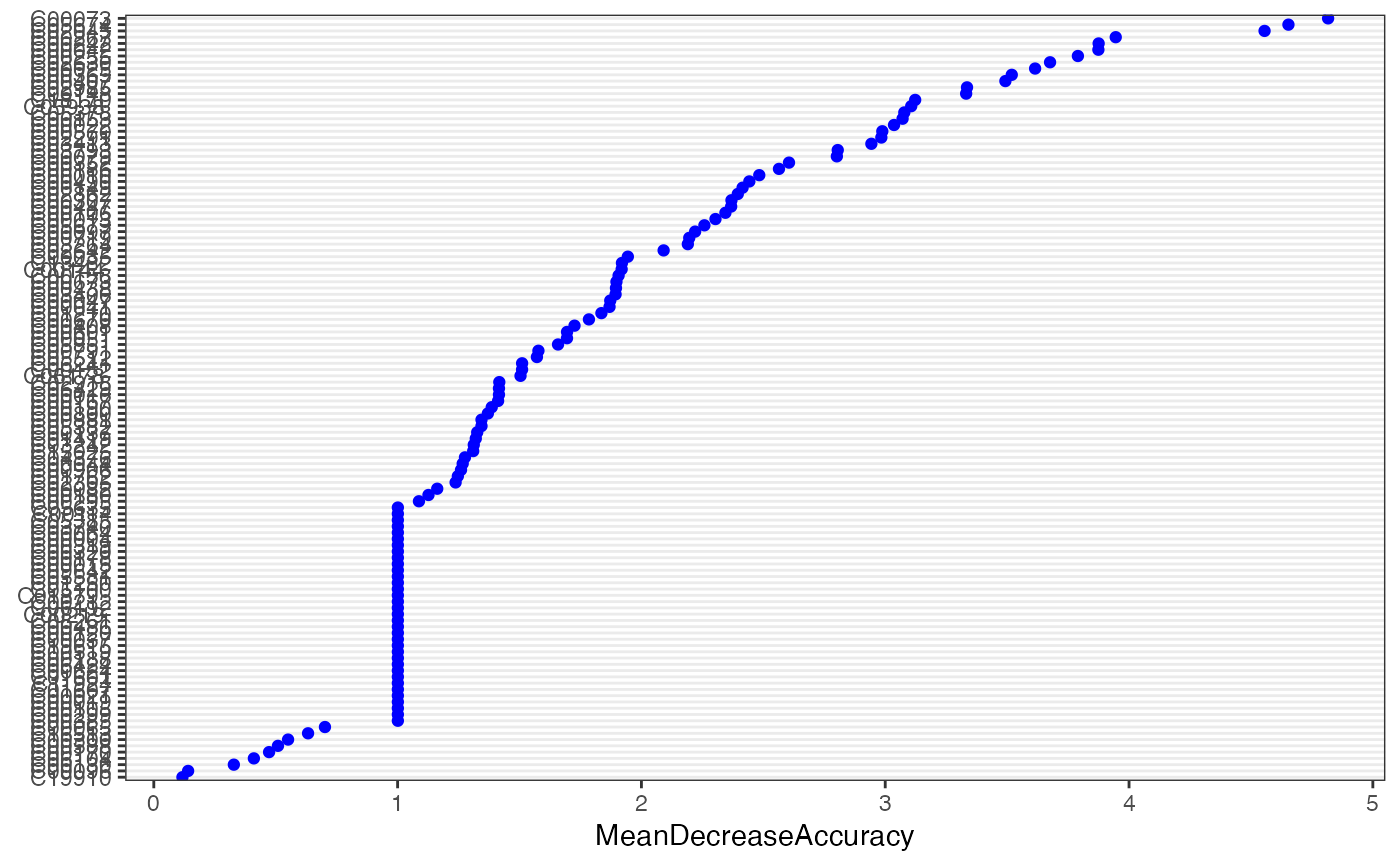 result_ML_RF$feature_result
#> # A tibble: 122 × 6
#> normal tumor MeanDecreaseAccuracy MeanDecreaseGini names raw
#> <dbl> <dbl> <dbl> <dbl> <chr> <fct>
#> 1 4.58 4.50 4.82 0.576 C00073 C00073
#> 2 4.27 4.57 4.65 0.624 C05674 C05674
#> 3 4.01 4.41 4.56 0.597 C02045 C02045
#> 4 3.43 3.42 3.95 0.380 C00267 C00267
#> 5 3.77 3.41 3.88 0.366 C00242 C00242
#> 6 3.83 3.17 3.87 0.483 C00042 C00042
#> 7 2.82 3.64 3.79 0.524 C00255 C00255
#> 8 3.72 2.37 3.68 0.529 C02630 C02630
#> 9 3.83 3.06 3.61 0.373 C00025 C00025
#> 10 3.86 2.57 3.52 0.435 C00365 C00365
#> # ℹ 112 more rows
result_ML_RF$feature_result
#> # A tibble: 122 × 6
#> normal tumor MeanDecreaseAccuracy MeanDecreaseGini names raw
#> <dbl> <dbl> <dbl> <dbl> <chr> <fct>
#> 1 4.58 4.50 4.82 0.576 C00073 C00073
#> 2 4.27 4.57 4.65 0.624 C05674 C05674
#> 3 4.01 4.41 4.56 0.597 C02045 C02045
#> 4 3.43 3.42 3.95 0.380 C00267 C00267
#> 5 3.77 3.41 3.88 0.366 C00242 C00242
#> 6 3.83 3.17 3.87 0.483 C00042 C00042
#> 7 2.82 3.64 3.79 0.524 C00255 C00255
#> 8 3.72 2.37 3.68 0.529 C02630 C02630
#> 9 3.83 3.06 3.61 0.373 C00025 C00025
#> 10 3.86 2.57 3.52 0.435 C00365 C00365
#> # ℹ 112 more rows