Metabolism-related subnetwork analysis
Source:vignettes/02-Subnetwork_analysis.Rmd
02-Subnetwork_analysis.RmdDifferential Information
The case study by Liao et al. (2022) integrated metabolic and transcriptional analysis to reveal elevated pyrimidine metabolism and glutaminolysis in TNBC among 31 breast tumors, and classified them into two clusters. We utilize MNet to identify a set of features whose activities significantly differ between these two clusters. This result will hopefully hint at some specific biological activities that are pathologically altered in tumoral samples.
Subnetwork identified by dnet
Input data must include the “name” column, with “p_value” required and “logFC” optional. If “logFC” exists, it dictates color; otherwise, use blue for metabolites and red for genes.
## meta_dat is the metabolic data of the 31 samples
## gene_dat is the transcriptional data of the 31 samples
## group is the group information of the 31 samples
## mlimma is the function of Differential Metabolite analysis by limma
diff_meta <- mlimma(meta_dat, group)
head(diff_meta)## # A tibble: 6 × 8
## logFC AveExpr t P.Value adj.P.Val B logP name
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <chr>
## 1 2.86 22.6 9.25 1.52e-10 0.0000000332 14.1 7.48 C02045
## 2 2.44 26.2 7.80 6.83e- 9 0.000000748 10.4 6.13 C00267
## 3 -1.82 27.1 -6.80 1.10e- 7 0.00000622 7.64 5.21 C00073
## 4 -3.78 20.9 -6.79 1.14e- 7 0.00000622 7.61 5.21 C05674
## 5 -2.20 21.4 -6.58 2.07e- 7 0.00000907 7.02 5.04 C00255
## 6 -2.37 21.6 -6.45 2.98e- 7 0.0000109 6.66 4.96 C00242## # A tibble: 6 × 8
## logFC AveExpr t P.Value adj.P.Val B logP name
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <chr>
## 1 6.92 23.1 11.3 1.07e-12 0.0000000190 17.9 7.72 APH1B
## 2 21.4 23.4 10.6 5.69e-12 0.0000000506 16.4 7.30 GFRA1
## 3 -5.76 24.4 -10.2 1.67e-11 0.0000000937 15.5 7.03 RFC4
## 4 10.5 14.8 10.1 2.11e-11 0.0000000937 15.3 7.03 FAM47E
## 5 4.02 5.65 9.83 3.69e-11 0.000000120 14.8 6.92 FAM87B
## 6 -10.8 20.6 -9.79 4.07e-11 0.000000120 14.7 6.92 ORC6
## change the name in diff_meta 'P.Value' to 'p_value'
names(diff_meta)[4] <- "p_value"
## filter the differential metabolites
diff_metabolite <- diff_meta %>%
filter(adj.P.Val < 0.01) %>%
filter(abs(logFC) > 1)
## change the name in diff_gene 'P.Value' to 'p_value'
names(diff_gene)[4] <- "p_value"
## filter the differential expression genes
diff_gene1 <- diff_gene %>%
filter(adj.P.Val < 0.01) %>%
filter(abs(logFC) > 1)
# dir.create("result_v0131")
# png("result_v0131/subnetwork_important.png",width = 8, height = 7, units = 'in', res = 200)
## identify the core metabolism-related subnetwork
a <- sNETlyser(diff_meta, diff_gene, nsize = 100)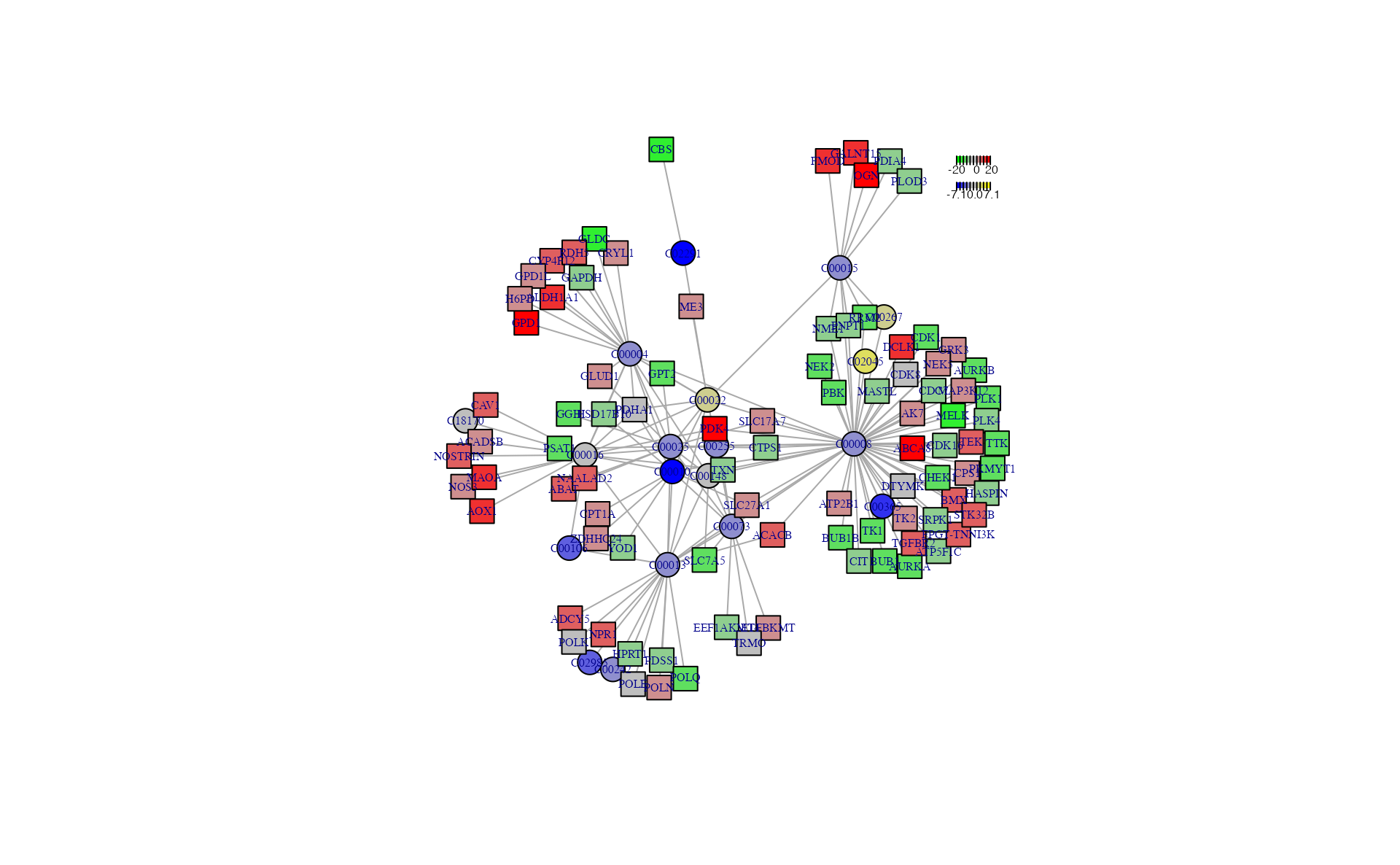
a## $node_result
## name type logFC AveExpr t p_value adj.P.Val
## 1 C00004 metabolite -2.32 21.727047 -4.043754 3.105194e-04 9.855615e-04
## 2 C00008 metabolite -1.61 26.915728 -2.558933 1.544157e-02 2.749352e-02
## 3 C00010 metabolite -6.77 17.135068 -6.060921 9.173483e-07 1.912691e-05
## 4 C00013 metabolite -2.26 26.993427 -5.547469 4.064386e-06 5.235885e-05
## 5 C00015 metabolite -2.35 25.255687 -3.450511 1.594418e-03 4.013535e-03
## 6 C00016 metabolite -0.77 19.350309 -1.675330 1.036357e-01 1.473781e-01
## 7 C00025 metabolite -1.52 30.828161 -5.098364 1.500590e-05 1.060094e-04
## 8 C00365 metabolite -4.49 19.519903 -5.795534 1.977561e-06 2.706787e-05
## 9 C02985 metabolite -3.00 22.074992 -6.328344 4.247004e-07 1.328706e-05
## 10 C00022 metabolite 1.44 24.204231 5.959762 1.228914e-06 2.070247e-05
## 11 C00106 metabolite -3.11 26.684736 -6.154394 7.005083e-07 1.704570e-05
## 12 C00255 metabolite -2.20 21.357147 -6.579475 2.070069e-07 9.066904e-06
## 13 C00148 metabolite -1.40 30.504922 -5.866104 1.611709e-06 2.353096e-05
## 14 C00267 metabolite 2.44 26.228537 7.801170 6.831988e-09 7.481027e-07
## 15 C00073 metabolite -1.82 27.121139 -6.800424 1.104723e-07 6.216787e-06
## 16 C00242 metabolite -2.37 21.589952 -6.451754 2.981604e-07 1.088285e-05
## 17 C02291 metabolite -7.05 19.394642 -6.002475 1.086113e-06 1.982156e-05
## 18 C02045 metabolite 2.86 22.643320 9.246113 1.517272e-10 3.322827e-08
## 19 C18170 metabolite -1.41 30.500751 -5.916654 1.392209e-06 2.177813e-05
## 20 ABCA8 gene 17.72 21.429976 7.911609 1.354128e-08 1.613957e-06
## 21 ACACB gene 10.75 26.910121 6.313422 4.516557e-07 1.626968e-05
## 22 ADCY5 gene 10.80 17.439272 6.437534 3.166774e-07 1.271965e-05
## 23 AK7 gene 5.04 9.457650 6.506762 3.524098e-07 1.345902e-05
## 24 ATP2B1 gene 4.47 29.300110 5.578654 3.764011e-06 7.754650e-05
## 25 ATP5F1C gene -4.98 35.514591 -6.094144 8.479140e-07 2.565265e-05
## 26 AURKA gene -10.91 22.465755 -7.947439 4.735183e-09 8.977915e-07
## 27 AURKB gene -11.36 19.110752 -8.407832 1.384197e-09 5.343902e-07
## 28 BMX gene 8.87 10.651736 6.995449 7.770982e-08 4.981215e-06
## 29 BUB1 gene -11.71 23.210846 -8.186199 2.494013e-09 6.920496e-07
## 30 BUB1B gene -10.30 22.881880 -6.346401 4.109495e-07 1.526789e-05
## 31 CDC7 gene -7.02 22.392840 -7.234139 3.347271e-08 2.817260e-06
## 32 CDK1 gene -11.01 26.430882 -7.396614 2.133124e-08 2.152395e-06
## 33 CDK16 gene -4.90 33.659519 -7.806509 6.935946e-09 1.080486e-06
## 34 CDK8 gene -3.66 22.779754 -5.693552 2.697715e-06 6.026254e-05
## 35 CHEK1 gene -8.41 22.346608 -7.024691 6.008033e-08 4.174576e-06
## 36 CIT gene -6.87 22.564460 -6.156151 7.093574e-07 2.273913e-05
## 37 CPS1 gene 7.39 14.277831 5.992796 1.135548e-06 3.141152e-05
## 38 CTPS1 gene -7.25 27.931682 -6.441867 3.127827e-07 1.268198e-05
## 39 DCLK1 gene 14.01 23.498349 6.533492 2.800217e-07 1.161478e-05
## 40 DTYMK gene -3.91 22.038522 -5.835299 1.789736e-06 4.401625e-05
## 41 FPGT-TNNI3K gene 8.35 12.342927 8.131239 4.696583e-09 8.977915e-07
## 42 GRK3 gene 4.84 27.363790 7.163480 7.235965e-08 4.782652e-06
## 43 HASPIN gene -7.20 14.494119 -7.243967 3.257007e-08 2.769621e-06
## 44 MAP3K12 gene 7.54 24.538204 6.106373 8.185818e-07 2.497800e-05
## 45 MASTL gene -7.12 25.792464 -9.594579 6.589670e-11 1.376351e-07
## 46 MELK gene -12.39 21.556778 -7.958982 4.589921e-09 8.977915e-07
## 47 NEK2 gene -11.86 20.658434 -7.053846 5.536721e-08 4.079943e-06
## 48 NEK5 gene 6.98 13.398510 6.675615 1.608072e-07 8.067159e-06
## 49 NME1 gene -5.39 29.622976 -7.305005 2.749100e-08 2.556087e-06
## 50 PBK gene -9.74 19.095824 -5.757435 2.242037e-06 5.189385e-05
## 51 PDK4 gene 19.91 25.564214 8.399442 1.415235e-09 5.347482e-07
## 52 PKMYT1 gene -10.18 20.944080 -7.284947 2.906475e-08 2.666337e-06
## 53 PLK1 gene -10.59 22.624463 -6.746873 1.314085e-07 7.108426e-06
## 54 PLK4 gene -7.96 21.251613 -6.669639 1.635564e-07 8.158984e-06
## 55 RRM2 gene -10.65 26.406588 -6.726438 1.392349e-07 7.359144e-06
## 56 SLC17A7 gene 6.80 9.548279 7.088822 6.012667e-08 4.174576e-06
## 57 SLC27A1 gene 6.12 24.168456 6.253613 5.361454e-07 1.852414e-05
## 58 SRPK1 gene -6.23 32.110850 -8.768319 5.384049e-10 3.328580e-07
## 59 STK32B gene 10.17 15.665397 6.881147 1.523235e-07 7.795716e-06
## 60 TEK gene 11.94 19.310413 6.680809 1.584555e-07 7.994348e-06
## 61 TGFBR2 gene 9.47 35.070723 7.617670 1.161018e-08 1.509869e-06
## 62 TK1 gene -9.85 24.293472 -6.584032 2.085826e-07 9.720281e-06
## 63 TK2 gene 4.61 26.796432 7.262399 3.094332e-08 2.707007e-06
## 64 TTK gene -10.76 20.572338 -7.604942 1.202218e-08 1.514198e-06
## 65 ALDH1A1 gene 14.49 22.024927 5.706844 2.595805e-06 5.805906e-05
## 66 CRYL1 gene 4.60 20.724954 5.517996 4.488220e-06 8.846425e-05
## 67 CYP4F12 gene 10.04 12.190344 6.226939 1.027564e-06 2.981700e-05
## 68 GAPDH gene -6.76 50.561018 -5.514507 4.533881e-06 8.897630e-05
## 69 GLDC gene -14.89 17.466434 -6.837514 1.017142e-07 6.041279e-06
## 70 GLUD1 gene 4.87 33.841440 6.017013 1.058943e-06 3.023436e-05
## 71 GPD1 gene 16.24 19.173724 6.350855 4.057439e-07 1.521514e-05
## 72 GPD1L gene 6.03 27.548533 6.904397 8.423838e-08 5.286181e-06
## 73 H6PD gene 5.57 32.217726 7.325256 2.598966e-08 2.442065e-06
## 74 HSD17B10 gene -4.14 28.572926 -6.104926 8.219978e-07 2.500034e-05
## 75 PDHA1 gene -3.76 31.935650 -5.765686 2.189113e-06 5.108602e-05
## 76 RDH5 gene 8.56 14.448947 5.989148 1.147564e-06 3.169453e-05
## 77 ME3 gene 5.02 21.964317 6.137682 1.124639e-06 3.137603e-05
## 78 NOS3 gene 5.68 18.890199 5.593991 3.600263e-06 7.469284e-05
## 79 TXN gene -4.94 33.816196 -6.696250 1.516682e-07 7.784614e-06
## 80 PNPT1 gene -4.56 28.953012 -5.529923 4.335548e-06 8.622061e-05
## 81 CPT1A gene 5.55 29.734599 6.104347 8.233680e-07 2.500034e-05
## 82 YOD1 gene -5.81 25.782081 -5.977330 1.187367e-06 3.264156e-05
## 83 ZDHHC24 gene 4.07 21.947734 5.812928 1.909381e-06 4.600908e-05
## 84 HPRT1 gene -4.73 26.038333 -5.533988 4.284716e-06 8.549694e-05
## 85 NPR1 gene 9.96 20.181792 7.180314 3.888535e-08 3.110653e-06
## 86 PDSS1 gene -6.37 18.246207 -7.266648 3.058015e-08 2.700543e-06
## 87 POLE gene -3.14 30.147049 -6.022212 1.043187e-06 3.010673e-05
## 88 POLK gene 2.87 26.124109 6.086482 8.668279e-07 2.600337e-05
## 89 POLN gene 7.10 11.987359 6.210572 6.066559e-07 2.010403e-05
## 90 POLQ gene -9.85 20.506744 -8.436628 1.282846e-09 5.177740e-07
## 91 FMOD gene 15.43 28.243500 7.096981 4.907142e-08 3.708338e-06
## 92 GALNT15 gene 12.32 18.290691 6.678378 2.613687e-07 1.120036e-05
## 93 OGN gene 18.20 22.806139 7.790483 9.023024e-09 1.281919e-06
## 94 PDIA4 gene -5.68 36.460830 -6.931848 7.797611e-08 4.981215e-06
## 95 PLOD3 gene -4.43 29.982526 -5.874824 1.596457e-06 4.021487e-05
## 96 ACADSB gene 7.89 28.574986 6.149912 7.221998e-07 2.306753e-05
## 97 AOX1 gene 14.53 18.984377 8.620853 1.032967e-09 4.703708e-07
## 98 CAV1 gene 9.26 32.804468 6.946964 7.473100e-08 4.915363e-06
## 99 MAOA gene 12.46 20.677373 7.184017 3.848607e-08 3.101698e-06
## 100 NOSTRIN gene 10.62 19.919745 7.268492 3.042382e-08 2.700543e-06
## 101 EEF1AKMT4 gene -7.15 17.236700 -7.610448 1.184217e-08 1.512986e-06
## 102 ETFBKMT gene 4.95 17.269675 6.043198 9.819487e-07 2.872887e-05
## 103 TRMO gene 3.29 20.426508 6.557063 2.252186e-07 1.017724e-05
## 104 GPT2 gene -8.56 28.350456 -5.995227 1.127614e-06 3.138760e-05
## 105 ABAT gene 10.38 21.786314 5.951486 1.279323e-06 3.431949e-05
## 106 GGH gene -8.08 24.429955 -6.676473 1.604163e-07 8.067159e-06
## 107 NAALAD2 gene 9.17 13.369770 7.240810 3.285726e-08 2.778629e-06
## 108 PSAT1 gene -11.58 22.681462 -7.655738 1.046140e-08 1.429108e-06
## 109 SLC7A5 gene -10.07 30.536982 -5.806475 1.945362e-06 4.658300e-05
## 110 CBS gene -12.49 19.845748 -5.793222 2.021413e-06 4.799235e-05
## B logP colors
## 1 -0.1284396 3.0063163 #8F8FCE
## 2 -3.8094665 1.5607697 #8F8FCE
## 3 5.5544799 4.7183551 #0000FF
## 4 4.0931288 4.2810099 #8F8FCE
## 5 -1.6936270 2.3964730 #8F8FCE
## 6 -5.4722678 0.8315672 #BEBEBE
## 7 2.8143804 3.9746554 #8F8FCE
## 8 4.7999437 4.5675459 #3030EF
## 9 6.3116406 4.8765713 #5F5FDF
## 10 5.2671639 4.6839779 #CECE8F
## 11 5.8195612 4.7683851 #5F5FDF
## 12 7.0186328 5.0425410 #8F8FCE
## 13 5.0008122 4.6283604 #BEBEBE
## 14 10.3759397 6.1260388 #CECE8F
## 15 7.6366728 5.2064340 #8F8FCE
## 16 6.6596209 4.9632572 #8F8FCE
## 17 5.3885292 4.7028621 #0000FF
## 18 14.1148491 7.4784923 #DFDF5F
## 19 5.1446045 4.6619794 #BEBEBE
## 20 9.4064440 5.7921081 #FF0000
## 21 6.3180451 4.7886209 #DF5F5F
## 22 6.6453036 4.8955247 #DF5F5F
## 23 6.5336480 4.8709865 #CE8F8F
## 24 4.3575662 4.1104378 #CE8F8F
## 25 5.7366528 4.5908678 #8FCE8F
## 26 10.4851053 6.0468245 #5FDF5F
## 27 11.5936062 6.2721416 #5FDF5F
## 28 7.9249493 5.3026647 #DF5F5F
## 29 11.0639486 6.1598628 #5FDF5F
## 30 6.4051392 4.8162209 #5FDF5F
## 31 8.7071806 5.5501731 #8FCE8F
## 32 9.1182958 5.6670780 #5FDF5F
## 33 10.1395031 5.9663807 #8FCE8F
## 34 4.6660836 4.2199526 #BEBEBE
## 35 8.1722065 5.3793876 #5FDF5F
## 36 5.9014425 4.6432262 #8FCE8F
## 37 5.4667196 4.5029110 #CE8F8F
## 38 6.6567039 4.8968129 #8FCE8F
## 39 6.7527009 4.9349889 #EF3030
## 40 5.0459289 4.3563869 #BEBEBE
## 41 10.4318173 6.0468245 #DF5F5F
## 42 7.9516134 5.3203312 #CE8F8F
## 43 8.7321481 5.5575797 #8FCE8F
## 44 5.7691751 4.6024424 #CE8F8F
## 45 14.2998757 6.8612709 #8FCE8F
## 46 10.5132846 6.0468245 #30EF30
## 47 8.2470016 5.3893459 #5FDF5F
## 48 7.2689283 5.0932794 #CE8F8F
## 49 8.8869226 5.5924244 #8FCE8F
## 50 4.8373894 4.2848841 #5FDF5F
## 51 11.5736912 6.2718507 #FF0000
## 52 8.8361150 5.5740850 #5FDF5F
## 53 7.4544376 5.1482266 #5FDF5F
## 54 7.2533459 5.0883639 #8FCE8F
## 55 7.4012950 5.1331727 #5FDF5F
## 56 8.1587543 5.3793876 #CE8F8F
## 57 6.1598543 4.7322620 #CE8F8F
## 58 12.4389931 6.4777410 #8FCE8F
## 59 7.2806506 5.1081440 #DF5F5F
## 60 7.2824695 5.0972170 #DF5F5F
## 61 9.6719418 5.8210607 #DF5F5F
## 62 7.0297155 5.0123212 #5FDF5F
## 63 8.7789365 5.5675107 #CE8F8F
## 64 9.6402482 5.8198173 #5FDF5F
## 65 4.7017421 4.2361300 #EF3030
## 66 4.1945145 4.0532322 #CE8F8F
## 67 5.5367471 4.5255360 #DF5F5F
## 68 4.1851343 4.0507257 #8FCE8F
## 69 7.6895939 5.2188711 #30EF30
## 70 5.5312821 4.5194992 #CE8F8F
## 71 6.4168928 4.8177241 #FF0000
## 72 7.8625171 5.2768579 #CE8F8F
## 73 8.9381665 5.6122428 #CE8F8F
## 74 5.7653283 4.6020541 #8FCE8F
## 75 4.8595030 4.2916979 #BEBEBE
## 76 5.4569887 4.4990156 #DF5F5F
## 77 5.4656404 4.5034020 #CE8F8F
## 78 4.3987731 4.1267210 #CE8F8F
## 79 7.3227038 5.1087629 #8FCE8F
## 80 4.2265846 4.0643889 #8FCE8F
## 81 5.7637897 4.6020541 #CE8F8F
## 82 5.4254657 4.4862291 #8FCE8F
## 83 4.9860454 4.3371565 #CE8F8F
## 84 4.2375132 4.0680494 #8FCE8F
## 85 8.5702288 5.5071485 #DF5F5F
## 86 8.7897157 5.5685489 #8FCE8F
## 87 5.5451378 4.5213364 #BEBEBE
## 88 5.7162718 4.5849703 #BEBEBE
## 89 6.0458297 4.6967168 #CE8F8F
## 90 11.6618718 6.2858598 #5FDF5F
## 91 8.3574706 5.4308207 #EF3030
## 92 6.7922162 4.9507682 #EF3030
## 93 9.8780484 5.8921394 #FF0000
## 94 7.9333387 5.3026647 #8FCE8F
## 95 5.1516663 4.3956134 #8FCE8F
## 96 5.8848743 4.6369989 #CE8F8F
## 97 11.8192399 6.3275597 #EF3030
## 98 7.9723006 5.3084444 #DF5F5F
## 99 8.5796621 5.5084004 #EF3030
## 100 8.7943953 5.5685489 #DF5F5F
## 101 9.6539608 5.8201650 #8FCE8F
## 102 5.6010497 4.5416814 #CE8F8F
## 103 6.9591088 4.9923699 #BEBEBE
## 104 5.4732016 4.5032419 #5FDF5F
## 105 5.3564951 4.4644592 #DF5F5F
## 106 7.2711649 5.0932794 #5FDF5F
## 107 8.7241302 5.5561694 #DF5F5F
## 108 9.7666045 5.8449349 #5FDF5F
## 109 4.9687674 4.3317725 #5FDF5F
## 110 4.9332749 4.3188280 #30EF30
##
## $edge_result
## X1 X2
## 1 C00004 C00008
## 2 C00004 C00010
## 3 C00008 C00010
## 4 C00008 C00013
## 5 C00010 C00013
## 6 C00008 C00015
## 7 C00004 C00016
## 8 C00010 C00016
## 9 C00013 C00016
## 10 C00004 C00025
## 11 C00008 C00025
## 12 C00010 C00025
## 13 C00013 C00025
## 14 C00008 C00365
## 15 C00013 C02985
## 16 C00004 C00022
## 17 C00008 C00022
## 18 C00010 C00022
## 19 C00013 C00022
## 20 C00015 C00022
## 21 C00016 C00022
## 22 C00025 C00022
## 23 C00013 C00106
## 24 C00016 C00106
## 25 C00008 C00255
## 26 C00016 C00255
## 27 C00004 C00148
## 28 C00008 C00148
## 29 C00013 C00148
## 30 C00025 C00148
## 31 C00008 C00267
## 32 C00015 C00267
## 33 C00008 C00073
## 34 C00010 C00073
## 35 C00013 C00073
## 36 C00025 C00073
## 37 C00022 C00073
## 38 C00148 C00073
## 39 C00013 C00242
## 40 C00022 C02291
## 41 C00008 C02045
## 42 C00016 C18170
## 43 C00008 ABCA8
## 44 C00008 ACACB
## 45 C00013 ACACB
## 46 C00013 ADCY5
## 47 C00008 AK7
## 48 C00008 ATP2B1
## 49 C00008 ATP5F1C
## 50 C00008 AURKA
## 51 C00008 AURKB
## 52 C00008 BMX
## 53 C00008 BUB1
## 54 C00008 BUB1B
## 55 C00008 CDC7
## 56 C00008 CDK1
## 57 C00008 CDK16
## 58 C00008 CDK8
## 59 C00008 CHEK1
## 60 C00008 CIT
## 61 C00008 CPS1
## 62 C00008 CTPS1
## 63 C00025 CTPS1
## 64 C00008 DCLK1
## 65 C00008 DTYMK
## 66 C00365 DTYMK
## 67 C00008 FPGT-TNNI3K
## 68 C00008 GRK3
## 69 C00008 HASPIN
## 70 C00008 MAP3K12
## 71 C00008 MASTL
## 72 C00008 MELK
## 73 C00008 NEK2
## 74 C00008 NEK5
## 75 C00008 NME1
## 76 C00015 NME1
## 77 C00008 PBK
## 78 C00008 PDK4
## 79 C00016 PDK4
## 80 C00008 PKMYT1
## 81 C00008 PLK1
## 82 C00008 PLK4
## 83 C00008 RRM2
## 84 C00015 RRM2
## 85 C00008 SLC17A7
## 86 C00025 SLC17A7
## 87 C00008 SLC27A1
## 88 C00010 SLC27A1
## 89 C00013 SLC27A1
## 90 C00008 SRPK1
## 91 C00008 STK32B
## 92 C00008 TEK
## 93 C00008 TGFBR2
## 94 C00008 TK1
## 95 C00365 TK1
## 96 C00008 TK2
## 97 C00365 TK2
## 98 C00008 TTK
## 99 C00004 ALDH1A1
## 100 C00004 CRYL1
## 101 C00004 CYP4F12
## 102 C00004 GAPDH
## 103 C00004 GLDC
## 104 C00004 GLUD1
## 105 C00025 GLUD1
## 106 C00004 GPD1
## 107 C00004 GPD1L
## 108 C00004 H6PD
## 109 C00004 HSD17B10
## 110 C00010 HSD17B10
## 111 C00016 HSD17B10
## 112 C00004 PDHA1
## 113 C00010 PDHA1
## 114 C00016 PDHA1
## 115 C00022 PDHA1
## 116 C00004 RDH5
## 117 C00022 ME3
## 118 C00016 NOS3
## 119 C00008 TXN
## 120 C00016 TXN
## 121 C00073 TXN
## 122 C00008 PNPT1
## 123 C00015 PNPT1
## 124 C00010 CPT1A
## 125 C00010 YOD1
## 126 C00010 ZDHHC24
## 127 C00013 HPRT1
## 128 C00242 HPRT1
## 129 C00013 NPR1
## 130 C00013 PDSS1
## 131 C00013 POLE
## 132 C00013 POLK
## 133 C00013 POLN
## 134 C00013 POLQ
## 135 C00015 FMOD
## 136 C00015 GALNT15
## 137 C00015 OGN
## 138 C00015 PDIA4
## 139 C00015 PLOD3
## 140 C00016 ACADSB
## 141 C00016 AOX1
## 142 C00016 CAV1
## 143 C00016 MAOA
## 144 C00016 NOSTRIN
## 145 C00073 EEF1AKMT4
## 146 C00073 ETFBKMT
## 147 C00073 TRMO
## 148 C00025 GPT2
## 149 C00022 GPT2
## 150 C00025 ABAT
## 151 C00025 GGH
## 152 C00025 NAALAD2
## 153 C00025 PSAT1
## 154 C00148 SLC7A5
## 155 C00073 SLC7A5
## 156 C02291 CBS
# dev.off()
# write.table(
# a$node_result,
# "result_v0131/subnetwork_important_node.txt",
# quote = F,
# row.names = F,
# sep = "\t"
# )
# write.table(
# a$edge_result,
# "result_v0131/subnetwork_important_edge.txt",
# quote = F,
# row.names = F,
# sep = "\t"
# )Subnetwork extraction of interested metabolites and genes
Input data requires the “name” column but excludes the “p_value” column.
# png("result_v0131/subnetwork_interested.png",width = 8, height = 7, units = 'in', res = 200)
## get 500 differential expression gene
a <- sNETlyser(diff_metabolite[, 8], diff_gene1[1:500, 8])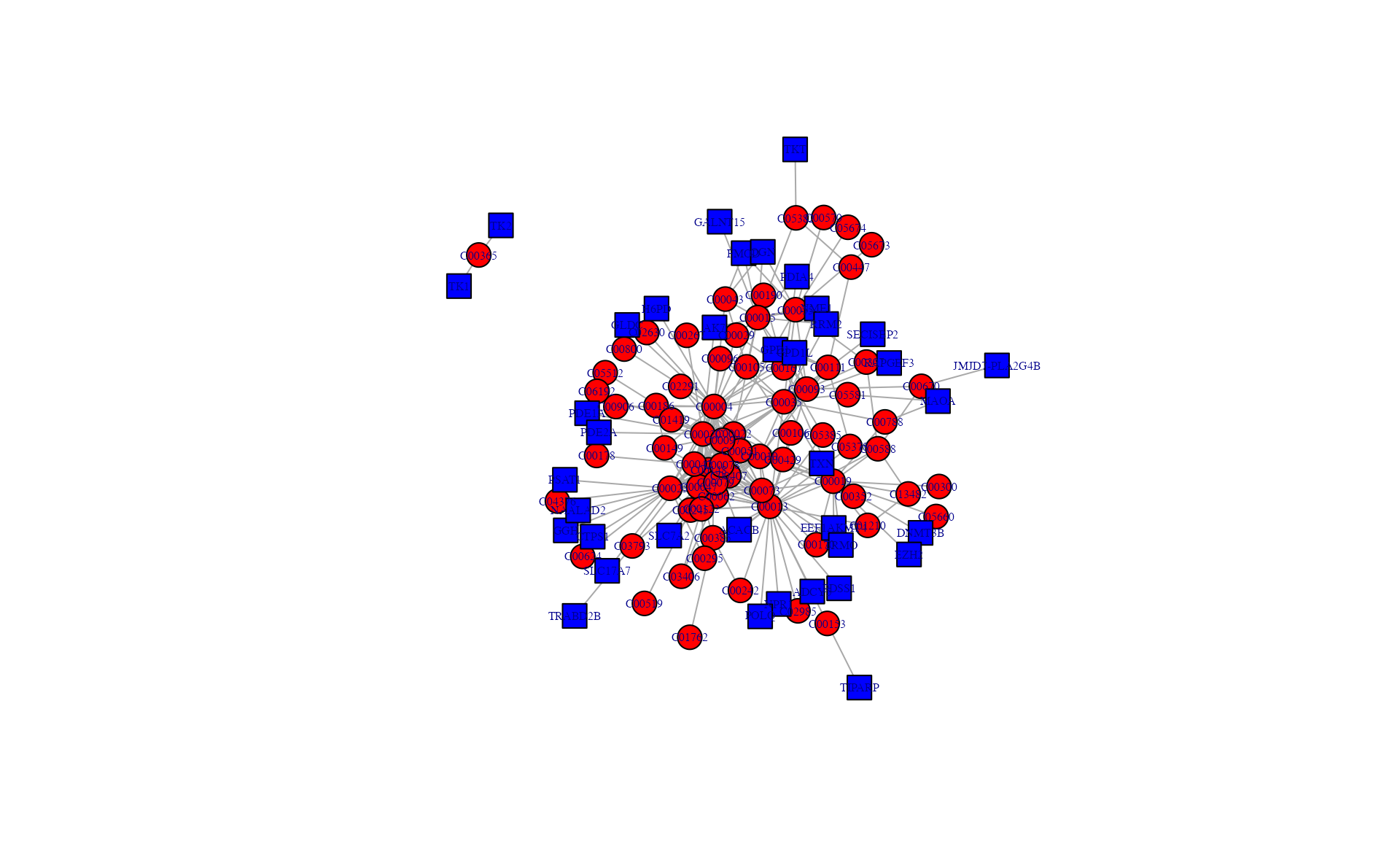
a## X1 X2
## 1 C00010 C00004
## 2 C00020 C00004
## 3 C00022 C00004
## 4 C00025 C00004
## 5 C00029 C00004
## 6 C00051 C00004
## 7 C00062 C00004
## 8 C00093 C00004
## 9 C00097 C00004
## 10 C00148 C00004
## 11 C00149 C00004
## 12 C00167 C00004
## 13 C00186 C00004
## 14 C00385 C00004
## 15 C00429 C00004
## 16 C00800 C00004
## 17 C05581 C00004
## 18 C00010 C00013
## 19 C00010 C00019
## 20 C00010 C00020
## 21 C00010 C00025
## 22 C00010 C00035
## 23 C00010 C00047
## 24 C00010 C00051
## 25 C00010 C00073
## 26 C00010 C00093
## 27 C00010 C00097
## 28 C00010 C00111
## 29 C00010 C00245
## 30 C00010 C00352
## 31 C00010 C00407
## 32 C00013 C00042
## 33 C00013 C00073
## 34 C00097 C00013
## 35 C00013 C00106
## 36 C00013 C00153
## 37 C00013 C00242
## 38 C00013 C00245
## 39 C00013 C00295
## 40 C00385 C00013
## 41 C00429 C00013
## 42 C00013 C00588
## 43 C00013 C02985
## 44 C00013 C05378
## 45 C00029 C00015
## 46 C00043 C00015
## 47 C00055 C00015
## 48 C00105 C00015
## 49 C00167 C00015
## 50 C00190 C00015
## 51 C05382 C00015
## 52 C00013 C00019
## 53 C00020 C00019
## 54 C00051 C00019
## 55 C00019 C00073
## 56 C00167 C00019
## 57 C00019 C00170
## 58 C00019 C00300
## 59 C00019 C00588
## 60 C00019 C00788
## 61 C00019 C01210
## 62 C00019 C05660
## 63 C00019 C13482
## 64 C00020 C00013
## 65 C00020 C00015
## 66 C00020 C00042
## 67 C00020 C00051
## 68 C00020 C00097
## 69 C00020 C00245
## 70 C00010 C00022
## 71 C00022 C00013
## 72 C00022 C00015
## 73 C00022 C00019
## 74 C00020 C00022
## 75 C00022 C00035
## 76 C00022 C00073
## 77 C00022 C00097
## 78 C00022 C00149
## 79 C00022 C00186
## 80 C00025 C00013
## 81 C00020 C00025
## 82 C00022 C00025
## 83 C00025 C00035
## 84 C00025 C00042
## 85 C00025 C00051
## 86 C00025 C00073
## 87 C00025 C00079
## 88 C00025 C00097
## 89 C00025 C00148
## 90 C00025 C00295
## 91 C00025 C00407
## 92 C00025 C00624
## 93 C00025 C04376
## 94 C00029 C00105
## 95 C00029 C00167
## 96 C00013 C00035
## 97 C00020 C00035
## 98 C00035 C00042
## 99 C00035 C00055
## 100 C00035 C00096
## 101 C00035 C00105
## 102 C00042 C00004
## 103 C00097 C00042
## 104 C00042 C00122
## 105 C00148 C00042
## 106 C00042 C03793
## 107 C00043 C00004
## 108 C00043 C00105
## 109 C00047 C00004
## 110 C00013 C00047
## 111 C00020 C00047
## 112 C00025 C00047
## 113 C00047 C00042
## 114 C00062 C00047
## 115 C00047 C00073
## 116 C00047 C00078
## 117 C00047 C00079
## 118 C00097 C00047
## 119 C00148 C00047
## 120 C00047 C00407
## 121 C00051 C00097
## 122 C00051 C01419
## 123 C00093 C00055
## 124 C00097 C00055
## 125 C00055 C00307
## 126 C00055 C00570
## 127 C00010 C00062
## 128 C00062 C00013
## 129 C00020 C00062
## 130 C00062 C00042
## 131 C00062 C00073
## 132 C00062 C00078
## 133 C00062 C00079
## 134 C00062 C00097
## 135 C00062 C00148
## 136 C00062 C00407
## 137 C00062 C03406
## 138 C00020 C00073
## 139 C00073 C00078
## 140 C00073 C00079
## 141 C00097 C00073
## 142 C00148 C00073
## 143 C00073 C00407
## 144 C00078 C00004
## 145 C00013 C00078
## 146 C00020 C00078
## 147 C00022 C00078
## 148 C00025 C00078
## 149 C00079 C00078
## 150 C00097 C00078
## 151 C00148 C00078
## 152 C00407 C00078
## 153 C00079 C00004
## 154 C00010 C00079
## 155 C00013 C00079
## 156 C00020 C00079
## 157 C00097 C00079
## 158 C00148 C00079
## 159 C00407 C00079
## 160 C00093 C00013
## 161 C00093 C00015
## 162 C00093 C00111
## 163 C00093 C00670
## 164 C00096 C00004
## 165 C00097 C00148
## 166 C00097 C00407
## 167 C00097 C01419
## 168 C00097 C02291
## 169 C00042 C00105
## 170 C00106 C00105
## 171 C00167 C00105
## 172 C00105 C00190
## 173 C00429 C00106
## 174 C00013 C00122
## 175 C00020 C00122
## 176 C00022 C00122
## 177 C00062 C00122
## 178 C00149 C00122
## 179 C00295 C00122
## 180 C00122 C03406
## 181 C00148 C00013
## 182 C00020 C00148
## 183 C00148 C00407
## 184 C00149 C00042
## 185 C00149 C00186
## 186 C00167 C00190
## 187 C00073 C00170
## 188 C00042 C00178
## 189 C00178 C00906
## 190 C00385 C00242
## 191 C00025 C00245
## 192 C00245 C00042
## 193 C00097 C00245
## 194 C00267 C00015
## 195 C00020 C00267
## 196 C00093 C00307
## 197 C00588 C00307
## 198 C00385 C00042
## 199 C00385 C01762
## 200 C00407 C00004
## 201 C00013 C00407
## 202 C00020 C00407
## 203 C00407 C00042
## 204 C00111 C00447
## 205 C05382 C00447
## 206 C00245 C00519
## 207 C00588 C00670
## 208 C00588 C13482
## 209 C00035 C00788
## 210 C00906 C00004
## 211 C00013 C01210
## 212 C01210 C13482
## 213 C00022 C02291
## 214 C02630 C00004
## 215 C00047 C03793
## 216 C00111 C05378
## 217 C00013 C05385
## 218 C00167 C05385
## 219 C00020 C05512
## 220 C00055 C05673
## 221 C00055 C05674
## 222 C00020 C06192
## 223 C00004 GLDC
## 224 C00004 GPD1
## 225 C00004 GPD1L
## 226 C00004 H6PD
## 227 C00013 ACACB
## 228 C00013 ADCY5
## 229 C00013 NPR1
## 230 C00013 PDSS1
## 231 C00013 POLQ
## 232 C00015 FMOD
## 233 C00015 GALNT15
## 234 C00015 NME1
## 235 C00015 OGN
## 236 C00015 PDIA4
## 237 C00015 RRM2
## 238 C00019 DNMT3B
## 239 C00019 EEF1AKMT4
## 240 C00019 EZH2
## 241 C00019 TRMO
## 242 C00020 ACACB
## 243 C00020 AK7
## 244 C00020 PDE1A
## 245 C00020 PDE2A
## 246 C00025 CTPS1
## 247 C00025 GGH
## 248 C00025 NAALAD2
## 249 C00025 PSAT1
## 250 C00025 SLC17A7
## 251 C00035 NME1
## 252 C00035 RAPGEF3
## 253 C00035 RRM2
## 254 C00035 SECISBP2
## 255 C00035 TXN
## 256 C00043 FMOD
## 257 C00043 OGN
## 258 C00047 SLC7A2
## 259 C00055 AK7
## 260 C00055 FMOD
## 261 C00055 OGN
## 262 C00062 SLC7A2
## 263 C00073 EEF1AKMT4
## 264 C00073 TRMO
## 265 C00073 TXN
## 266 C00079 SLC7A2
## 267 C00093 GPD1
## 268 C00093 GPD1L
## 269 C00111 GPD1
## 270 C00111 GPD1L
## 271 C00153 TIPARP
## 272 C00167 PDIA4
## 273 C00365 TK1
## 274 C00365 TK2
## 275 C00670 JMJD7-PLA2G4B
## 276 C00788 MAOA
## 277 C03793 TRABD2B
## 278 C05382 TKT
## 279 C05581 MAOA
# dev.off()
# write.table(a,"result_v0131/subnetwork_interested_edge.txt",quote=F,row.names=F,sep="\t")Subnetwork extraction using correlation
# png("result_v0131/subnetwork_correlation.png",width = 8, height = 7, units = 'in', res = 200)
## extract the correlation subnetwork
a <- pNetCor(meta_dat, gene_dat, cor_threshold = 0.95)## [1] "Starting correlation calculation"
## [1] "If the data is large, it will take some minutes"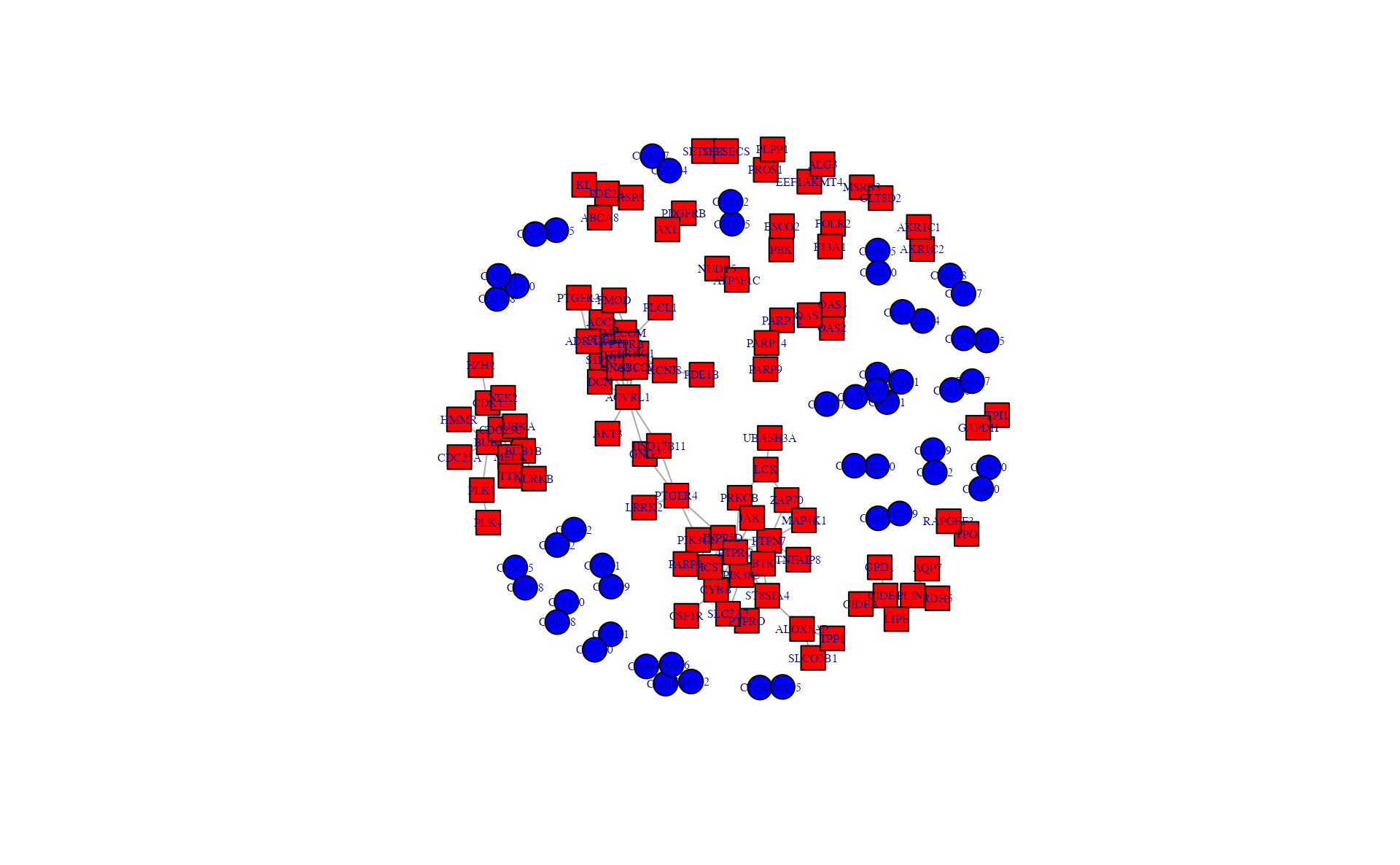
a## $result
## name1 name2 cor_result p type1 type2
## 1 C05100 C00408 0.9938503 0.000000e+00 metabolite metabolite
## 2 C00408 C00064 0.9938415 0.000000e+00 metabolite metabolite
## 3 C06178 C00047 0.9980774 0.000000e+00 metabolite metabolite
## 4 C02220 C01042 0.9745498 0.000000e+00 metabolite metabolite
## 5 C00881 C00380 0.9543875 0.000000e+00 metabolite metabolite
## 6 C02630 C00490 0.9519425 2.220446e-16 metabolite metabolite
## 7 C00632 C00299 0.9843097 0.000000e+00 metabolite metabolite
## 8 C05100 C00064 0.9999999 0.000000e+00 metabolite metabolite
## 9 C05635 C00328 0.9997317 0.000000e+00 metabolite metabolite
## 10 C18170 C00148 0.9999757 0.000000e+00 metabolite metabolite
## 11 C06192 C00096 0.9519796 2.220446e-16 metabolite metabolite
## 12 C06192 C00190 0.9570487 0.000000e+00 metabolite metabolite
## 13 C02045 C00267 0.9521105 2.220446e-16 metabolite metabolite
## 14 C00075 C00002 0.9575343 0.000000e+00 metabolite metabolite
## 15 C05851 C00180 0.9991263 0.000000e+00 metabolite metabolite
## 16 C00180 C00079 0.9543253 0.000000e+00 metabolite metabolite
## 17 C00180 C00082 0.9968227 0.000000e+00 metabolite metabolite
## 18 C00601 C00180 0.9911016 0.000000e+00 metabolite metabolite
## 19 C00475 C00120 0.9808235 0.000000e+00 metabolite metabolite
## 20 C00570 C00307 0.9655488 0.000000e+00 metabolite metabolite
## 21 C05851 C00079 0.9554133 0.000000e+00 metabolite metabolite
## 22 C05851 C00082 0.9963136 0.000000e+00 metabolite metabolite
## 23 C05851 C00601 0.9919686 0.000000e+00 metabolite metabolite
## 24 C00149 C00122 0.9960786 0.000000e+00 metabolite metabolite
## 25 C00035 C00015 0.9683562 0.000000e+00 metabolite metabolite
## 26 C00190 C00096 0.9505742 4.440892e-16 metabolite metabolite
## 27 C00096 C00043 0.9574429 0.000000e+00 metabolite metabolite
## 28 C01419 C00051 0.9698552 0.000000e+00 metabolite metabolite
## 29 C00144 C00105 0.9568950 0.000000e+00 metabolite metabolite
## 30 C00387 C00294 0.9619177 0.000000e+00 metabolite metabolite
## 31 C05938 C00025 0.9999788 0.000000e+00 metabolite metabolite
## 32 C02642 C00152 0.9999987 0.000000e+00 metabolite metabolite
## 33 C00407 C00079 0.9825118 0.000000e+00 metabolite metabolite
## 34 C00082 C00079 0.9524477 0.000000e+00 metabolite metabolite
## 35 C00601 C00082 0.9902250 0.000000e+00 metabolite metabolite
## 36 C00190 C00043 0.9645821 0.000000e+00 metabolite metabolite
## 37 PDE2A ABCA8 0.9521911 2.220446e-14 gene gene
## 38 ACVRL1 ABCC9 0.9611130 0.000000e+00 gene gene
## 39 KCNJ8 ABCC9 0.9718499 0.000000e+00 gene gene
## 40 MECOM ABCC9 0.9695464 0.000000e+00 gene gene
## 41 PDE1A ABCC9 0.9599045 2.220446e-16 gene gene
## 42 PRKG1 ABCC9 0.9612267 0.000000e+00 gene gene
## 43 PTPRB ABCC9 0.9703811 0.000000e+00 gene gene
## 44 S1PR1 ABCC9 0.9528181 1.776357e-15 gene gene
## 45 TEK ABCC9 0.9596725 2.220446e-16 gene gene
## 46 AKT3 ACVRL1 0.9512478 2.220446e-16 gene gene
## 47 DCN ACVRL1 0.9641382 0.000000e+00 gene gene
## 48 GNG11 ACVRL1 0.9702326 0.000000e+00 gene gene
## 49 GNG2 ACVRL1 0.9548637 2.220446e-16 gene gene
## 50 HSD17B11 ACVRL1 0.9510524 2.664535e-15 gene gene
## 51 PRKG1 ACVRL1 0.9513762 2.220446e-16 gene gene
## 52 PTPRB ACVRL1 0.9684628 0.000000e+00 gene gene
## 53 S1PR1 ACVRL1 0.9702329 0.000000e+00 gene gene
## 54 TEK ACVRL1 0.9536665 0.000000e+00 gene gene
## 55 AOC3 ADRA2A 0.9529137 5.329071e-15 gene gene
## 56 DCN ADRA2A 0.9510561 8.881784e-15 gene gene
## 57 GNG11 ADRA2A 0.9558340 2.442491e-15 gene gene
## 58 PDE1A ADRA2A 0.9665477 2.220446e-16 gene gene
## 59 PTGER3 ADRA2A 0.9515055 7.993606e-15 gene gene
## 60 PTPRB ADRA2A 0.9591008 8.881784e-16 gene gene
## 61 S1PR1 ADRA2A 0.9674976 0.000000e+00 gene gene
## 62 TEK ADRA2A 0.9658895 0.000000e+00 gene gene
## 63 AKR1C2 AKR1C1 0.9508092 2.220446e-16 gene gene
## 64 EEF1AKMT4 ALG3 0.9617165 0.000000e+00 gene gene
## 65 SLCO2B1 ALOX5AP 0.9540727 3.996803e-15 gene gene
## 66 ST8SIA4 ALOX5AP 0.9622097 4.440892e-16 gene gene
## 67 TPP1 ALOX5AP 0.9572490 1.554312e-15 gene gene
## 68 FMOD AOC3 0.9505416 4.440892e-16 gene gene
## 69 MECOM AOC3 0.9591528 0.000000e+00 gene gene
## 70 PDE1A AOC3 0.9612698 0.000000e+00 gene gene
## 71 PTPRB AOC3 0.9685499 0.000000e+00 gene gene
## 72 S1PR1 AOC3 0.9608990 0.000000e+00 gene gene
## 73 TEK AOC3 0.9504502 4.440892e-16 gene gene
## 74 PLIN1 AQP7 0.9645697 1.252332e-13 gene gene
## 75 PDE2A ASPA 0.9581644 1.332268e-15 gene gene
## 76 NUDT5 ATP5F1C 0.9537685 0.000000e+00 gene gene
## 77 BUB1 AURKA 0.9691365 0.000000e+00 gene gene
## 78 BUB1B AURKA 0.9542584 0.000000e+00 gene gene
## 79 CDC25C AURKA 0.9757903 0.000000e+00 gene gene
## 80 CDK1 AURKA 0.9615361 0.000000e+00 gene gene
## 81 MELK AURKA 0.9553172 0.000000e+00 gene gene
## 82 BUB1B AURKB 0.9608279 0.000000e+00 gene gene
## 83 MELK AURKB 0.9569659 0.000000e+00 gene gene
## 84 TTK AURKB 0.9586561 0.000000e+00 gene gene
## 85 PDGFRB AXL 0.9507983 2.220446e-16 gene gene
## 86 INPP5D BTK 0.9568919 0.000000e+00 gene gene
## 87 PIK3R5 BTK 0.9535169 0.000000e+00 gene gene
## 88 PTPN7 BTK 0.9666415 0.000000e+00 gene gene
## 89 PTPRC BTK 0.9606287 0.000000e+00 gene gene
## 90 ST8SIA4 BTK 0.9536160 1.332268e-15 gene gene
## 91 TNFAIP8 BTK 0.9595595 0.000000e+00 gene gene
## 92 BUB1B BUB1 0.9624841 0.000000e+00 gene gene
## 93 CDC25A BUB1 0.9551371 0.000000e+00 gene gene
## 94 CDC25C BUB1 0.9711373 0.000000e+00 gene gene
## 95 CDK1 BUB1 0.9568210 0.000000e+00 gene gene
## 96 HMMR BUB1 0.9667073 0.000000e+00 gene gene
## 97 MELK BUB1 0.9703135 0.000000e+00 gene gene
## 98 NEK2 BUB1 0.9587695 0.000000e+00 gene gene
## 99 PLK1 BUB1 0.9691279 0.000000e+00 gene gene
## 100 TTK BUB1 0.9608439 0.000000e+00 gene gene
## 101 CDC25C BUB1B 0.9627357 0.000000e+00 gene gene
## 102 MELK BUB1B 0.9666373 0.000000e+00 gene gene
## 103 TTK BUB1B 0.9675330 0.000000e+00 gene gene
## 104 CDK1 CDC25C 0.9536671 0.000000e+00 gene gene
## 105 MELK CDC25C 0.9674048 0.000000e+00 gene gene
## 106 EZH2 CDK1 0.9532571 0.000000e+00 gene gene
## 107 NEK2 CDK1 0.9555746 0.000000e+00 gene gene
## 108 CIDEC CIDEA 0.9631204 4.005130e-11 gene gene
## 109 GPD1 CIDEC 0.9552145 4.867884e-12 gene gene
## 110 LIPE CIDEC 0.9813491 8.881784e-16 gene gene
## 111 PLIN1 CIDEC 0.9735118 2.753353e-14 gene gene
## 112 CYBB CSF1R 0.9642722 0.000000e+00 gene gene
## 113 PIK3CG CYBB 0.9531043 4.440892e-16 gene gene
## 114 PIK3R5 CYBB 0.9534256 0.000000e+00 gene gene
## 115 PTPRC CYBB 0.9618130 0.000000e+00 gene gene
## 116 PTPRO CYBB 0.9641765 0.000000e+00 gene gene
## 117 SLC7A7 CYBB 0.9685405 0.000000e+00 gene gene
## 118 GNG11 DCN 0.9606695 4.440892e-16 gene gene
## 119 PRKG1 DCN 0.9512348 2.664535e-15 gene gene
## 120 PBK ESCO2 0.9611330 0.000000e+00 gene gene
## 121 FOLR2 F13A1 0.9539469 1.614264e-13 gene gene
## 122 PRKG1 FMOD 0.9508807 2.220446e-16 gene gene
## 123 PTGER3 FMOD 0.9542998 0.000000e+00 gene gene
## 124 TPI1 GAPDH 0.9594477 0.000000e+00 gene gene
## 125 MSRB3 GLT8D2 0.9703943 0.000000e+00 gene gene
## 126 KCNJ8 GNG11 0.9530706 5.329071e-15 gene gene
## 127 MECOM GNG11 0.9554520 2.664535e-15 gene gene
## 128 PDE1A GNG11 0.9559116 8.215650e-15 gene gene
## 129 PRKG1 GNG11 0.9557364 2.442491e-15 gene gene
## 130 PTPRB GNG11 0.9660518 0.000000e+00 gene gene
## 131 S1PR1 GNG11 0.9631806 2.220446e-16 gene gene
## 132 TEK GNG11 0.9615761 4.440892e-16 gene gene
## 133 HSD17B11 GNG2 0.9527556 1.776357e-15 gene gene
## 134 PTGER4 GNG2 0.9682377 0.000000e+00 gene gene
## 135 INPP5D HCST 0.9547074 8.881784e-16 gene gene
## 136 PIK3R5 HCST 0.9567257 4.440892e-16 gene gene
## 137 PTGER4 HSD17B11 0.9591484 8.881784e-16 gene gene
## 138 PIK3CG INPP5D 0.9659720 0.000000e+00 gene gene
## 139 PIK3R5 INPP5D 0.9597248 0.000000e+00 gene gene
## 140 PTGER4 INPP5D 0.9565467 0.000000e+00 gene gene
## 141 PTPN7 INPP5D 0.9599870 0.000000e+00 gene gene
## 142 PTPRC INPP5D 0.9698189 0.000000e+00 gene gene
## 143 PTPRC JAK3 0.9541770 0.000000e+00 gene gene
## 144 PDE1B KCNJ8 0.9535535 0.000000e+00 gene gene
## 145 PRKG1 KCNJ8 0.9616117 0.000000e+00 gene gene
## 146 PDE2A KL 0.9570194 1.776357e-15 gene gene
## 147 PRKCB LCK 0.9662961 0.000000e+00 gene gene
## 148 UBASH3A LCK 0.9530990 6.696865e-13 gene gene
## 149 ZAP70 LCK 0.9573878 0.000000e+00 gene gene
## 150 PLIN1 LIPE 0.9628021 0.000000e+00 gene gene
## 151 PTGER4 LRRK2 0.9694866 0.000000e+00 gene gene
## 152 PTPN7 MAP4K1 0.9751826 0.000000e+00 gene gene
## 153 ZAP70 MAP4K1 0.9612888 0.000000e+00 gene gene
## 154 PDE1A MECOM 0.9670994 0.000000e+00 gene gene
## 155 PRKG1 MECOM 0.9620268 0.000000e+00 gene gene
## 156 PTPRB MECOM 0.9766969 0.000000e+00 gene gene
## 157 S1PR1 MECOM 0.9647745 0.000000e+00 gene gene
## 158 TEK MECOM 0.9656955 0.000000e+00 gene gene
## 159 TTK MELK 0.9677622 0.000000e+00 gene gene
## 160 OAS2 OAS1 0.9809053 0.000000e+00 gene gene
## 161 OAS3 OAS1 0.9658351 0.000000e+00 gene gene
## 162 PARP12 OAS1 0.9543994 0.000000e+00 gene gene
## 163 OAS3 OAS2 0.9700924 0.000000e+00 gene gene
## 164 PARP14 PARP12 0.9591643 0.000000e+00 gene gene
## 165 PARP9 PARP14 0.9616310 0.000000e+00 gene gene
## 166 PTPRC PARP8 0.9618607 0.000000e+00 gene gene
## 167 PTPRB PDE1A 0.9657455 0.000000e+00 gene gene
## 168 S1PR1 PDE1A 0.9677138 0.000000e+00 gene gene
## 169 TEK PDE1A 0.9675572 0.000000e+00 gene gene
## 170 PTGER4 PIK3CG 0.9533792 1.332268e-15 gene gene
## 171 PTPRC PIK3CG 0.9611947 0.000000e+00 gene gene
## 172 PTPN7 PIK3R5 0.9506781 4.440892e-16 gene gene
## 173 PTPRC PIK3R5 0.9788964 0.000000e+00 gene gene
## 174 SLC7A7 PIK3R5 0.9557844 0.000000e+00 gene gene
## 175 ST8SIA4 PIK3R5 0.9706706 0.000000e+00 gene gene
## 176 PTPRB PLCL1 0.9550180 2.220446e-16 gene gene
## 177 RDH5 PLIN1 0.9500174 1.110223e-15 gene gene
## 178 PLK4 PLK1 0.9668259 0.000000e+00 gene gene
## 179 PROS1 PLPP1 0.9513904 2.442491e-15 gene gene
## 180 PTPRC PRKCB 0.9511774 8.881784e-16 gene gene
## 181 PTPRB PRKG1 0.9622456 0.000000e+00 gene gene
## 182 TEK PRKG1 0.9512029 2.220446e-16 gene gene
## 183 PTPRC PTPN7 0.9649503 0.000000e+00 gene gene
## 184 TNFAIP8 PTPN7 0.9550097 0.000000e+00 gene gene
## 185 ZAP70 PTPN7 0.9506275 4.440892e-16 gene gene
## 186 S1PR1 PTPRB 0.9827064 0.000000e+00 gene gene
## 187 TEK PTPRB 0.9729261 0.000000e+00 gene gene
## 188 ST8SIA4 PTPRC 0.9616076 0.000000e+00 gene gene
## 189 SLC7A7 PTPRO 0.9599202 2.220446e-16 gene gene
## 190 ST8SIA4 PTPRO 0.9554691 8.881784e-16 gene gene
## 191 TPO RAPGEF3 0.9506282 8.881784e-16 gene gene
## 192 TEK S1PR1 0.9775779 0.000000e+00 gene gene
## 193 SETDB2 SEPSECS 0.9587409 0.000000e+00 gene gene
##
## $p
## NULL
# dev.off()
# write.table(
# a$result,
# "result_v0131/subnetwork_correlation.txt",
# quote = F,
# row.names = F,
# sep = "\t"
# )