MNet Manual
2025-03-20
1 OVERVIEW
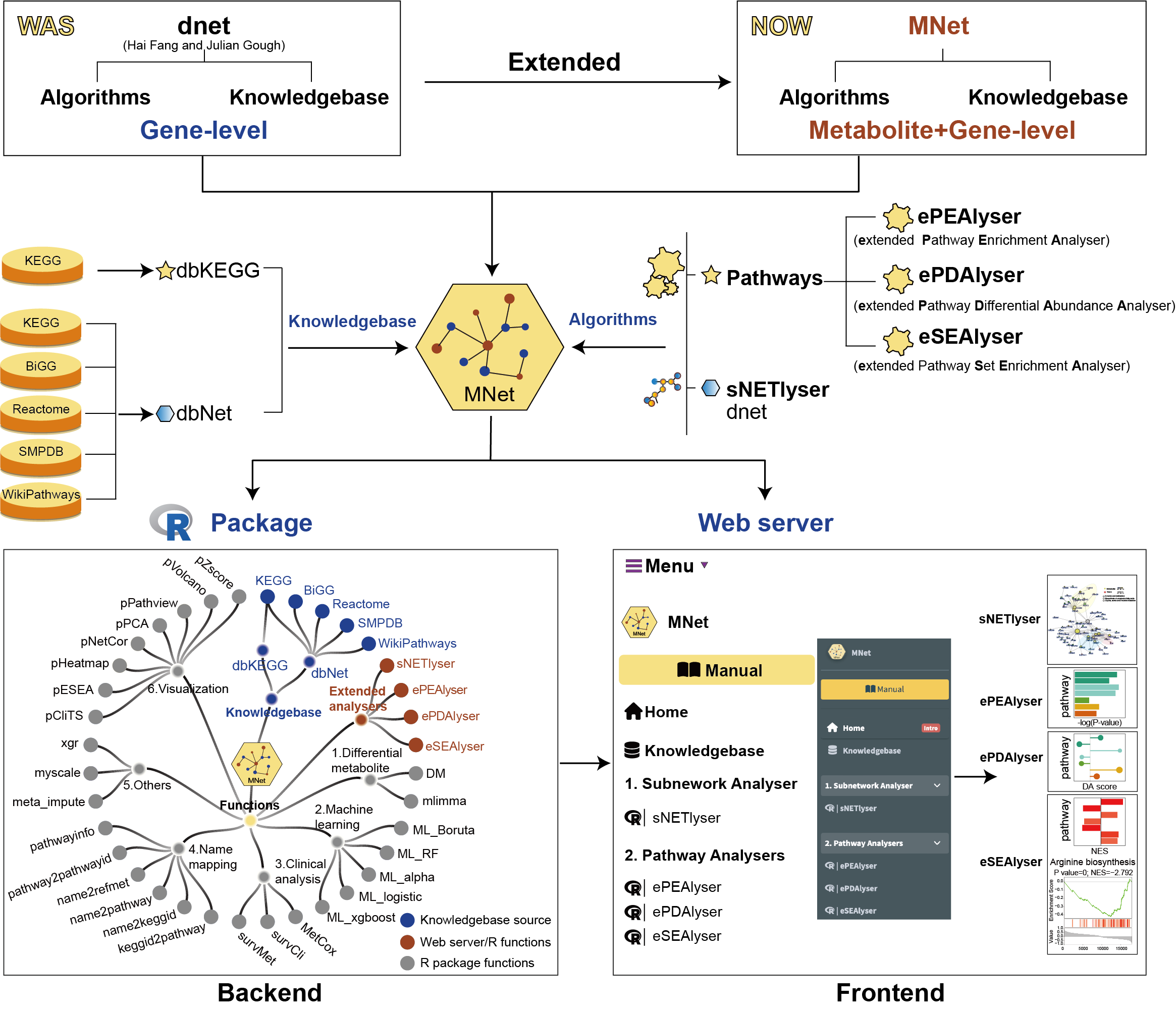
MNet features a built-in knowledgebase, dbMNet which is manually curated and provides information on metabolomics, including almost all currently available human metabolic interactions (gene-metabolite and metabolite-metabolite). Grounded in this knowledgebase, MNet offers one subnetwork analyser (sNETlyser) and three extended pathway analysers (ePEAlyser, ePDAlyser, and eSEAlyser) for user-input metabolomic and transcriptomic data.
Motivation
Advance since any previous publication (if relevant):
Integration of metabolomic and transcriptomic data is vital for pathway-centric, systems-biology understandings of disease (Nucleic Acids Research 2022 and Communications Biology 2023). Existing pathway knowledgebases, including but not limited to BiGG (Nucleic Acids Research 2020), KEGG (Nucleic Acids Research 2023), Reactome (Nucleic Acids Research 2024), SMPDB (Nucleic Acids Research 2014), and WikiPathways (Nucleic Acids Research 2024), provide metabolic pathway information on genes and metabolites. However, these databases lack the comprehensive integration of both metabolites and genes necessary for downstream pathway and subnetwork analyses, thereby limiting the exploration of potential therapeutic targets from a metabolomic perspective.
Our well-established algorithm/tool called ‘dnet’ and its related software(Genome Medicine 2014, Genome Medicine 2016 and Nature Genetics 2019) have received nearly 400 citations over time (AS OF October 2024 according to Google Scholar). They excel at identifying core subnetwork using prior knowledge but has been limited to genomic or transcriptomic challenges, without extending its application to metabolomics. In this aspect, MNet supports pathway-centric, network-driven analyses, enabling by the compilation of the dbMNet knowledgebase, which includes dbKEGG and dbNet.
The dbKEGG facilitates KEGG metabolic pathway-based extended pathway analysis to identify dysregulated metabolic pathways involving both metabolites and genes.
The dbNet enhances metabolism-related subnetwork analysis by leveraging gene-metabolite and metabolite-metabolite information to identify subnetwork that best explain the input data.
Browser compatibility
| MacOS (Big Sur) | Windows (10) | |
|---|---|---|
| Safari | 17.4.1 | N/A |
| Edge | N/A | 108.0.1462.54 |
| Chrome | 108.0.5359.124 | 108.0.5359.124 |